S.G. Nadeem, S.A. Qasmi, F. Afaque, M. Saleem and S.T. Hakim
Abstract
The objective of this study was to determine and compare thein vitro antibacterial susceptibility of 1008 Pseudomonas aeruginosa isolates obtained from 2800 clinical specimens received at a large hospital setting at Karachi, Pakistan between January 2008 and September 2008. Despite the widespread availability of antibiotics, it remains the most common bacterial infection in the humans. A total of 2800 clinical specimens were analysed for isolation and identification using standard isolation techniques mentioned in ASM’s Clinical Microbiology Manual[1]. Finally, 1008 found to be significant with Pseudomonas aeruginosa (36%), and subjected to antibiotic susceptibility testing in accordance with Kirby and Bauer disc diffusion method [2] and CLSI/NCCLS guidelines 2003 & 2007[8,9] , The isolated pathogens showed resistant to Amikacin (08%), Ceftriaxone(15%), Cefotaxime (16%), Sulzone (Cefapeozone+Sulbactum) (07%), Meropenam (08%), Ciprofloxacin (11%), and Fosfomycin (18%).
The results showed considerable variability in the size of zone of inhibition depending on which antibiotic was used. This study also reveals that resistance is developing to Imipenam , and Pseudomonas aeruginosa still remains an important cause of nosocomial infections. Keywords: Pseudomonas aeruginosa, Kirby and Bauer disc diffusion method, resistance
|
INTRODUCTION:
The genus pseudomonas are Gram-negative, aerobic, rod-shaped bacterium with unipolar motility,[1]contains more than 140 species, most of which are saprophytic. More than 25 species of pseudomonas are associated with humans [2]. Most pseudomonads known to cause disease in humans are associated with opportunistic infections. These include Ps. aeruginosa, Ps. fluorescens, Ps. putida, Ps. cepacia, Ps. stutzeri, Ps. maltophilia, and Ps. putrefaciens. Only two species, Ps. mallei and Ps. pseudomallei, produce specific human diseases: glanders and melioidosis. Ps. aeruginosa and Ps. maltophilia account for approximately 80 percent of pseudomonads recovered from clinical specimens [1,4].
Because of the frequency with which it is involved in human disease, Pseudomonas. aeruginosa has received the most attention. It is a ubiquitous free-living bacterium and is found in most moist environments. Although it seldom causes disease in healthy individuals, it is a major threat to hospitalised and immunocompromised patients, particularly those with serious underlying diseases such as cancer and burns [5]. The high mortality associated with these infections is due to a combination of weakened host defenses, bacterial resistance to antibiotics, and the production of extracellular bacterial enzymes and toxins [6].
Pseudomonas aeruginosa is a leading gram negative pathogen that causes nosocomial infections, accounting for 20% of pneumonia and 16% of urinary tract infections according to recent data from national nosocomial infection surveillance system [7]. According to the CDC, the overall incidence of Pseudomonas aeruginosa infections in U.S. hospitals averages about 0.4 percent (4 per 1000 discharges), and the bacterium is the fourth most commonly isolated nosocomial pathogen accounting for 10.1 percent of all hospital-acquired infections[9].
Resistance of this notorious bacterium to commonly used antimicrobial agents is becoming an increasing clinical problem and a recognised public health threat because there are limited number of antimicrobial agents including the antipseudomonal penicillins, cephalosporins, carbapenems, aminoglycosides and fluoroquinolones with reliable activity against it [11]. It has intrinsic resistance to many antimicrobial agents and only a few antimicrobial agents show potent antibacterial activity against this bacterium. The emergence of multidrug resistance (MDR) Pseudomonas aeruginosa has became a serious problem [12]. There are several mechanisms which may contribute to the antimicrobial resistance among Pseudomonas aeruginosa including the production of chromosomally encoded Amy C B-lactamases [13]. Hypermutable strains of Pseudomonas aeruginosa with defects in themethyl directed mismatch repair (MMR) system are also being frequently isolated from the lungs of cystic fibrosis (CF) patients [13].
MATERIALS AND METHODS:
Samples collection: For this study, a total of 1008 clinical isolates of Pseudomonas aeruginosa, were isolated from 2800 different clinical specimens including; urine (n= 905), ear swabs (n= 496), eye swabs (n=26), fluids (n= 31), pus swabs (n= 342), HVS (n= 157), and sputum (n= 843) received at the microbiology section of Burgor Anklesaria Hospital’s pathological laboratory between January 2008 and September 2008.
Primary isolation of test strains: For the primary isolation of test culture specimens were inoculated on routine culture media including CLED agar (Merck, Germany), EMB agar (Merck, Germany), MacConkey’s agar (Oxoid, UK), and Chocolate agar (Merck, Germany). Pigment production was interpreted on the basis of growth on Nutrient agar (Merck, Germany).
Control stain: ATCC Control strain of Pseudomonas aeruginosa(27853).
Spot tests: Selected colonies were further confirmed by spot tests including; Gram’s stain (Merck, Germany), Oxidase test (Oxoid, UK), Citrate utilisation test (Merck, Germany), and Urease tests (Merck, Germany) [1,4].
Sugar fermentation & IMVIC: Selected colonies were also subjected to Oxidative fermentation and IMVIC i.e. Indole, Methyl reductase test, Vogus prosekure test for confirmation of specie [1,4].
Antibacterial susceptibility testing: Antibacterial susceptibility testing of selected Pseudomonas aeruginosa species was done on Mueller Hinton agar (MHA) (Merck, Germany). To make bacterial suspensions, four to five colonies of pure growth from overnight cultures of test strains were transferred into a tube containing four to five millilitres of nutrient broth (Merck, Germany), and incubated at 37 °C to match the turbidity with McFarland’s index of 0.5 (usually 2-6 hours). Lawns of each bacterial suspension were made on MHA using sterile cotton swabs. Commercially available standard antibiotic discs of standardised concentrations (Oxoid, UK) (Amikacin, Ceftriaxone, Cefotaxime, Sulzone (Cefapeozone+Sulbactum), Meropenam, Ciprofloxacin, and Fosfomycin) were positioned at appropriate distances on the bacterial lawns and incubated at 37 °C for 24 hours. The growth inhibition zones were carefully measured with calipers and recorded according to the standard Kirby-Bauer disc diffusion method[2] and CLSI/NCCLS guidelines 2003 & 2007[8,9,13].
RESULTS:
This study was conducted on 2800 multiple type of clinical specimens received at Burgor Anklesaria Hospital’s pathological laboratory during January 2008 to September 2008. Out of these a total of 1008 clinical isolates were identified as Pseudomonas aeruginosa on the basis of gram’s stain and spot test reactions. Morphologically all of these isolates were gram negative, non sporing, capsulated, and motile short rods, produced typical grapes like odor of amino acetophenone and blue water soluble non fluorescent pigment pyocyanin.They were also positive for oxidase and citratase with variable ability to utilize urea agar. Of these1008 Ps. aeruginosa, 532 isolates were from male patients (504 adults and 28 children), and 476 isolates were from female patients (442 adults and 34 children) (Table 1).
Antibacterial susceptibility of seven selected antibiotics was determined against 1008 test strains of
Pseudomonas aeruginosa, using Kirby and Bauer disc diffusion method[2], against commercially available standardised antibiotic filter discs (Oxoid, UK). These strains were isolated from seven different categories of specimens including ear swabs, wound pus, urine, sputum, eye swab, fluids and high vaginal swab (HVS) (Table 2 & 3).
Another interesting observation was that a maximum number of test strains were isolated from urine i.e. 403 (40%). While, only 6 (0.6%) were isolated from eye swabs (Table 2). When susceptibility results were compared according to the age and sex, not a significant difference was observed (Table 3).
Out of a total of 504 isolates from male adults, 45 (9%) were resistant to Amikacin, 140 (28%) were resistant Ciprofloxacin, 185 (37%) were resistant to Cefotaxime, 174 (34%) were resistant to Ceftriaxone, 34 (7%) were resistant to Sulzone, 140 (28%) were resistant to Fosfornycin and 25 (6%) were resistant to Meropenam. Among 28 male children, the maximum resistance was observed to Ciprofloxacin (Table 3) out of 442 isolates from female adults 39 (9%) were resistant to Amikacin 84 (19%) were resistant to Ciprofloxacin, 78 (18%) were resistant to Cefotaxime, 151 (34%) were resistant to Ceftriaxone, 28 (6%) were resistant S ulzone (Cefapeozone+Sulbactum), 145 (33%) were resistant to Fosformycin and 11 (2%) were resistant to Meropenam. On the whole, the maximum resistance was observed from the male adults isolates against Cefotaxime (n=185, 37%) and in the case of isolates from the female adults to Ceftriaxone (n=151, 34%) .
Collectively, we can say that maximum resistance was observed when target cells were subjected to antimicrobial susceptibility testing against third generation Cephalosporins i.e. Ceftriaxone and Cefotaxime. The most effective antibiotic in the isolates from the male patients was Sulzone (Cefapeozone+Sulbactum) i.e. 465 (92%), while in the case of isolates from female patients it was Meropenam i.e. 414 (94%) (Table 3).
Table 1: Age and Gender wise distribution of clinical isolates of Pseudomonas aeruginosa
Total No. of samples N | 2800 n (%) |
Positive for Ps. aeruginosa | 1008 (36%) |
Male adult (>12 years) | 504 (50%) |
Male children (0-12 years) | 28 (3 %) |
Female adult (>12 years) | 442 (44%) |
Female children (0-12 years) | 34 (3%) |
Table 2: Resistance of Pseudomonas aeruginosa from different clinical specimens to antibiotics determined by Kirby-Bauer disc diffusion method
%=Percentage, n= Individual type of sample, N=Total number of sample, AK= Amikacin (R =>22 mm), CRO= Ceftriaxone (R=>21 mm), CTX=Cefotaxime (R=>22 mm), SCF= Sulzone (R= > 20 mm) (Cefapeozone+Sulbactum), MEM=Meropenam (R= >18 mm), CIP=Ciprofloxacin (R= > 21mm), FOS=Fosfomycin (R= > 18mm ), µgms= micro grams, S= sensitive
Table 3: Age and Gender wise sensitivity of Pseudomonas aeruginosa from different clinical specimens to antibiotics determined by Kirby-Bauer disc diffusion method
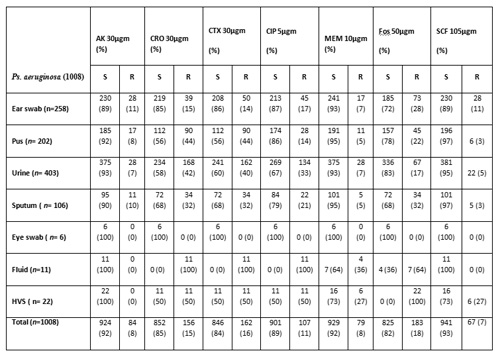
% =Percentage, n= Individual type of sample, N=Total number of sample, AK= Amikacin (R =>22 mm), CRO= Ceftriaxone (R=>21 mm), CTX=Cefotaxime (R=>22 mm), SCF= Sulzone (R= > 20 mm) (Cefapeozone+Sulbactum), MEM=Meropenam (R= >18 mm), CIP=Ciprofloxacin (R= > 21mm), FOS=Fosfomycin (R= > 18mm ), µgms= micro grams, S= sensitive
DISCUSSION:
Pseudomonas aeruginosa is a leading Gram-negative pathogen thatcauses nosocomial infections, accounting for 20% of pneumoniaand 16% of urinary tract infections according to recent datafrom the National Nosocomial Infections Surveillance System [1].
Optimisation of therapy against Pseudomonas aeruginosa starts with the initial empirical antibiotic choice. Surveillance data and hospital or unit antibiograms may inform this decision, although individualisation of the initial regimen on the basis of prior antibiotic use and prior isolation of resistant pathogens may be more important. Combinations of antibiotics are often required empirically, and "combination antibiograms" may need to be developed for this purpose. Preliminary data suggest that extending the time over which a dose of antipseudomonal beta-lactam antibiotics is infused may improve clinical outcomes; however, this idea remains to be confirmed in randomised trials. For example Moody et al in 1972 showed that some of the Pseudomonas speciesother than Pseudomonas. aeruginosa were resistant to a number of antibiotics.Among these were antibiotics that are in general use for P.aeruginosa infections. Such differences in antibiotic susceptibilities emphasise the necessity for careful speciation of this groupof microorganisms to assure proper epidemiological documentationof colonisation and infection, as well as to ensure therapywith an antimicrobial agent to which the organism is susceptiblein vitro. The role of direct susceptibility testing in aiding more rapid initiation of appropriate antibiotic therapy is also being studied. When identification and susceptibility testing is complete, the antibiotic regimen for infections due to Gram-negative pathogens can be "fine tuned." On some occasions, this fine tuning necessitates the introduction of "salvage" antibiotics, such as Colistin or Tigecycline; on others, it necessitates de-escalation and early termination of therapy. The lack of new antibiotic options against gram-negative pathogens underscores the need for optimisation of current therapies and prevention of the spread of these organisms.
In 2008 Javiya et al reported the highest number of Pseudomonas infections was found in urine, followed by pus and sputum. Pseudomonas species demonstrated marked resistance against monotherapy of penicillins, cephalosporins, fluoroquinolones, tetracyclines and macrolides. Only combination drugs like Ticarcillin + Clavulanic acid, Piperacillin + Tazobactum, Cefoperazone + Sulbactum, Cefotaxime + Sulbactum, Ceftriaxome + Sulbactum and monotherapy of Amikacin showed higher sensitivity to Pseudomonas infections; however, the maximum sensitivity was shown by the Carbapenems.
Our study was therefore carried out, using Kirby-Bauer method [2], to determine the antibiotic susceptibility patterns of Pseudomonas aeruginosa isolates from in-patients and out-patients attending the microbiology section of Burgor Anklesaria Hospital’s pathological laboratory between January 2008 and September 2008. The isolation rate of Pseudomonas aeruginosa in clinical specimens was found to be 36%, with the highest occurrence of 403 (40%) in urine samples followed by 258 (26%) occurrence in ear swabs. The susceptibility pattern showed that 85% were sensitive to Meropenam and 84% to Sulzone (Cefapeozone+Sulbactum). The isolates from the male patients showed almost equal resistance to all the antibiotics tested, as in case of isolates from the female patients, most especially Ceftriaxone and Cefotaxime. However, no consistent antibiotic susceptibility pattern could be established for this pathogenic bacterium based on sources.
Treatment of Pseudomonas aeruginosa is a challenge because resistance limits dramatically therapeutic options. In this review, we discuss data of in vitro susceptibility for the management of infections caused by Pseudomonas aeruginosa. Currently, published data from Pakistan are limited, and there are no such randomised clinical trials involving the treatment of infections caused by multidrug resistant Gram-negative rods. At present newer antimicrobial agents active against multidrug resistant bacteria like Pseudomonas aeruginosa are not available or under investigation.
CONCLUSION:
Antibiotic resistant organisms appear to be biologicallyfit and are capable of causing serious, life-threatening infectionsthat are difficult to manage because treatment options are limited.This increase in the prevalence of drug resistant pathogensis occurring at a time when the discovery and development ofnew anti-infective agents is slowing down dramatically.
The Pseudomonas aeruginosa species isolated from patients in the Microbiology section of Burgor Anklesaria Hospital’s pathological laboratory, Karachi, Pakistan were tested in vitro for antibacterial susceptibility of currently available and commonly prescribed drugs. Meropenam and Sulzone were the two antibiotics found to be the most susceptible against this pathogen. The emergence of multidrug resistant (MDR) Pseudomonas aeruginosa is a challenging clinical problem. This study investigated the pattern of antibiotic resistance to test antibiotics and helps us in determining the role of combination therapy in its management. The results of this study suggest that use of triple antimicrobial therapy (Meropenam, Sulzone and Amikacin) can be a useful alternative treatment for multidrug resistant (MDR) Pseudomonas aeruginosa infection in certain circumstances.
Acknowledgements
We are thankful to Dr. Dorab Anklesaria, Director, Burgor Anklesaria Hospital’s pathological laboratory, the administration of Jinnah University for Women, Karachi-Pakistan, and Mr. Syed Muhammad Humair Tayyab for their cooperation and support during this study.
Competing Interests
None Declared
Author Details
S G NADEEM, Department of Microbiology, Jinnah University for Women, Karachi-74600, Pakistan
S A QASMI, Liaquat College of Medicine and Dentistry, Karachi, Pakistan
F AFAQUE, Burgor Anklesaria Hospital’s Pathological Laboratory, Karachi, Pakistan
M SALEEM Burgor Anklesaria Hospital’s Pathological Laboratory, Karachi, Pakistan
S T HAKIM, Department of Microbiology, Jinnah University for Women, Karachi-74600, Pakistan
CORRESPONDENCE: SHAZIA TABASSUM HAKIM, Ph.D. Associate Professor of Microbiology & Virology,Department of Microbiology, Jinnah University for Women, Nazimabad, Karachi-74600, Pakistan
Email: Shaz2971@yahoo.com |
References
- Baron EJ, Jorgensen JH, Pfaller MA and Yolken RH (editors). 2003. ASM’s Manual of Clinical Microbiology (8th ed.). ASM Press, Washington. ISBN 1-55581-255-4.
- Ryan KJ, Ray CG (editors).2004. Sherris Medical Microbiology (4th ed.). McGraw Hill.ISBN 0-8385-8529-9.
- http://www.phageinternational.com/pathogens/pseudomonas.htm
- Bauer AW, Kirby WMM, Sherris JC and Turck M. 1966. Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol. 45: 493 – 96.
- Iglewski BH.1996. Pseudomonas. In: Baron's Medical Microbiology (Baron S et al, eds.)(4th ed.). Univ of Texas Medical Branch. ISBN 0-9631172-1-1.
- Anzai, et al. 2000, Jul. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol 50 (Pt 4): 1563–89.
- Worlitzsch D, Tarran R, Ulrich M, et al. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109 (3): 317–25
- Yakupogullari YL, Poirel S, Bernabeu A, Kizirgil, and P Nordmann. 2008. Multidrug-resistant Pseudomonas aeruginosa isolate co-expressing extended-spectrum β-lactamase PER-1 and metallo-β-lactamase VIM-2 from Turkey. J. Antimicrob.Chemo. 61(1): 221-222
- National Committee for Clinical Laboratory Standards.2003. NCCLS document M2-A8 volume 23, no. 1, Performance standards for antimicrobial disk susceptibility tests, approved standard, 8th ed. National Committee for Clinical Laboratory Standards, Villanova, PA.
- Clinical and Laboratory Standards Institute. 2007. M100-S17. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- Cooper M, Tavankar GR and Williams HD.2003. Regulation of expression of the cyanide-insensitive terminal oxidase in Pseudomonas aeruginosa. Microbiology 149 (Pt 5): 1275–84.
- Walkty AM, DeCorby K, Nichol JA, Karlowsky DJ, Hoban and Zhanel GG. 2008. In vitro activity of ceftobiprole against clinical isolates of Pseudomonas aeruginosa obtained from Canadian intensive care unit (ICU) patients as part of the CAN-ICU Study. J. Antimicrob.Chemo. 62(1): 206- 208.
- Kirikae T, Mizuguchi Y, and Arakawa Y. 2008. Investigation of isolation rates of Pseudomonas aeruginosa with and without multidrug resistance in medical facilities and clinical laboratories in Japan. J. Antimicrob.Chemo. 61(3): 612-615.
- Peter Z, and Borg MA. 1998. Antibiotic susceptibility patterns of local strains of Pseudomonas aeruginosa. Maltese Medical Journal. 10(1):11.
- Moody MR , Young VM, and Kenton DM.1972. In Vitro Antibiotic Susceptibility of Pseudomonads Other than Pseudomonas aeruginosa Recovered from Cancer Patients. Antimicrob Agents Chemother. 2(5): 344-349.
- Javiya VA et al. 2008. Antibiotic susceptibility patterns of Pseudomonas aeruginosa at a tertiary care hospital in Gujarat, India. Indian J Pharmacol. 40(5): 230-234.

The above article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.







